

|
Consuelo Junqueira Rodrigues*, Gina Camillo Silvestre*, Nilo Bozzini**, Aldo Junqueira Rodrigues Junior* |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Despite the benign status of uterine leiomyoma, chromosomal abnormalities involving the chromosome 7 have been described, suggesting the existence of tumor suppressor genes in this region. The shrinkage effect of GnRH agonist on leiomyoma is variable and may be related in part to an autonomous growth. The present study examine the association of the shrinkage effect of GnRH agonist treatment on uterine leiomyomas and microsatellite instability (MSI) and loss of hererozygosity (LOH) with D7S471, D7S496, D7S501, D7S515, D7S518 and D7S666 microsatellite marker. Twenty-nine nuliparous women with uterine leiomyomas, aging from 24 to 39 years old, submitted to US study of leiomyoma volume. They were treated with goserelin 3.6 mg every 28 days for 6 months. Twelve patients had leiomyoma reduction £ 36% and the other seventeen had reduction > 36%. All women were submitted to myomectomy. DNA was extracted from tumor paraffin-embedded tissue and matched peripheral blood. The PCR products were submitted to automated sequencer. We didn’t showed any association between MSI and leiomyoma shrinkage. There was a significant association between LOH in D7S515, D7S518 and D7S666 markers and high leiomyoma shrinkage. (Supported by FAPESP 01/14052-5).
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
Uterine leiomyoma is a benign neoplasm of the myometrium smooth muscle cells, responsive to steroid hormones. The development and growth of the leiomyomata result from a complex interaction of steroid hormones, progestins, growth factors, cytokines and somatic mutations. Both estrogen and progesterone are considered promoting factors, stimulating leiomyoma growth1-3. Leiomyomata development and growth involve many factors. The most important are family history and race. On the other hand, the risk of fibroids is lower in women with a higher number of term pregnancies and among users of oral contraceptive4,5. The leiomyoma volume seems to be associated with many cellular and extracellular factors, however there is a significant correlation between estrogen and progesterone receptors and leiomyoma growth6,7. Thus, patients that require a conservative treatment can receive gonadotropin releasing hormone (GnHR) agonist to induce estrogen depletion and consequently cause leiomyoma shrinkage8,9. Bozzini et al2 and Cirkel et al10 demonstrated a significant negative correlation between the number of estrogen receptor positive cells and the intensity of leiomyoma shrinkage in patients treated with GnRH. For its growth, the tumor requires two stages: initiation and promotion. Estrogen is considered the main promoter, with biochemical studies showing its role in leiomyoma growth stimulation, and it is present in higher concentration in the tumor than in the adjacent myometrium11. Many authors have demonstrated that the monthly administration of GnRH agonist significantly reduces the size of uterine leiomyomata12-15. Uterine leiomyoma patients receiving GnRH agonists present a varying percentage in uterus-leiomyoma shrinkage, demonstrating that the substance acts in a particular manner in each woman16. Bozzini et al2 observed varying uterus-leiomyoma shrinkage in nulliparous women treated with GnRH analog (goserelin). Friedman et al16 suggest that the variation in uterine leiomyoma shrinkage in patients receiving GnRH seems to be associated with the extracellular matrix reduction. Meanwhile, Rein et al11 proposed the possibility of a reduction in water and cellular content in the matrix. Kalir et al17 suggest that the polymerization of collagen fibrils after GnRH treatment can increase collagen concentration. Bozzini et al2 also demonstrated a collagen increase in leiomyomas that presented greater shrinkage after GnRH treatment. The hormonal effect variation on leiomyomata is partly associated with their autonomous growth. This autonomous growth can be associated with somatic mutations that induce chromosomal changes. Approximately 40% of the leiomyomata exhibit cytogenetic abnormalities, where the most frequent are translocations involving the 12q15 region and deletions of long arm of chromosome 718-24. The biological significance of these chromosomal abnormalities, involving growth or responsiveness to drugs, has not been definitively demonstrated yet. The most frequent leiomyoma cytogenetic change is the deletion of 7q22, found in approximately 35% of the studied cases. This same deletion of 7q22 has been verified in malignant neoplasms suggesting that a tumor suppressor gene could be located in this chromosomal region23. The analysis of chromosomal deletion by studies involving microsatellite loss of heterozygosity has shown better results than cytogenetic analysis, since it detects breakpoint deletions. Molecular genetic studies reveal a loss of heterozygosity in critical regions containing the leiomyomata suppressor gene. This loss is characterized by a change in regions of repeated nucleotides, known as microsatellites, defined by markers such as D7S518, D7S471, CULT1. Takahashi et al25 observed a loss of heterozygosity in the D7S491 region in leiomyomas that presented the greatest shrinkage in patients treated with GnRH. Despite the uncertainties regarding its real importance in the genome, the short tandem repeats (STRs) have been broadly used as tools in genetic mapping studies, population genetics, linkage analysis, evolution studies and genetic identification26,27. OBJECTIVE The objective of the present study was to verify the existence of an association between chromosome 7 abnormalities and uterus-leiomyoma shrinkage in nulliparous patients treated with GnRH analog.
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
3.1 CasuISTIC Twenty nine uterine leiomyoma patients were selected with ages ranging from 20 to 39 years, followed-up at the Hospital das Clínicas Gynecology Service from the University of São Paulo School of Medicine (HC-FMUSP). These patients were submitted to GnRH agonist treatment (goserelin) for a period of 6 months, at every 28 days, and underwent myomectomy on the sixth month after treatment. The uterus-leiomyoma volume was assessed by ultrasound, admission assessment, and at the end of the goserelin therapy. After six months using the GnRH analog (goserelin), ultrasound revealed different uterus-leiomyoma shrinkages, motivating the division of the group of patients into two subgroups. The division was based on the mean shrinkage percentage. Thus, subgroup 1 consisted of patients that had shrinkage equal to or below 36% (Table 1); subgroup 2 consisted of patients with shrinkage above 36% (Table 2). The genomic DNA was extracted from the patients’ paraffin-embedded leiomyoma block and peripheral blood, after free and informed consent. This research protocol was approved by the HC-FMUSP CAPPesq, number 066/02. 3.2 METHOD 3.2.1 Genomic DNA Extraction from Paraffin-Embedded Material The genomic DNA extraction from paraffin-embedded material was done as referred elsewhere28, using an extraction kit (WIZARD GENOMIC – PROMEGA). 3.2.2 Genomic DNA Extraction from Total Blood From each patient, 10 ml of peripheral blood was collected, transferred to an Eppendorf tube and submitted to the genomic DNA extraction method described by Miller et al29. After the genomic DNA had been extracted and quantified, it was submitted to integrity analysis in 2% agarose gel electrophoresis. 3.2.3 Polymerase Chain Reaction (PCR) Technique The forward and reverse primers were synthesized by Invitrogen do Brasil, with the forward primer labeled with fluorochromes according to the sequences below: D7S471: 5’CAACATATGCAAGGTGCCTA, 3’AGCAATTCCATAATAGCTGCT D7S496:5’AACAACAGTCAACCCACAAT, 3’TTTTGGTTTNTTATGGGTTATAGC D7S501: 5’CACCGTTGTGATGGCAGAG, 3’AGCAGTCTGCCTG GTAAGAAAT D7S515: 5’GGGAGTTACTACCCTCACTTAATG, 3’CTTTGCTGCCCAGTCC D7S518: 5’CAGTAGGCAGGGGTGG, 3’GTTGTCACACAGACACACCCC D7S666:5’GCCTTCTCAGCAAATTGAT, 3’CTCTTTCATTACCTCACA TATCAGG The PCR was done with the following solution: dNTPs (0.4 U dATP, 0.4U dCTP, 0.4U dGTP, 0.4U dTTP), 10X Buffer, Taq Platinum 1.5U Enzyme, Autoclaved Ultrapure Water, Forward and Reverse Primers (10pmol for peripheral blood and 50pmol for paraffin-embedded samples) and DNA. The success of the reaction was verified in 2% agarose gel with 0.1% ethidium bromide, in a horizontal gel electrophoresis and visualized under ultraviolet light. The PCR programs were similar for the paraffin-embedded material DNA and peripheral blood DNA regarding the number of cycles (34 cycles) and the extension time (10 minutes). The denaturing time varied, being 15’ for the paraffin embedded material and 5’ for the peripheral blood; the annealing time also varied, being 1’30" for the paraffin embedded material and 30" for the peripheral blood. The annealing temperatures were: 55oC for primers D7S496, D7S501 and D7S666; 53oC for primer D7S515 and 57oC for primers D7S471 and D7S518. 3.2.4 Instability Analysis and Microsatellite Loss of Heterozygosity The amplified fluorochrome-labeled products were submitted to MegaBace (Amersham Pharmacia Biotech, USA) automated sequencing. Microsatellite instability (MSI) was defined by changes in the amplified DNA segment length caused by insertion or deletion of repeated units of the tumor against normal tissue. The tumors with MSI in two or more markers were considered of high instability, those with one marker were considered of low instability and those that did not present MSI in any of the studied markers were considered stable. The loss of heterozygosity was assessed in the informative cases and was considered present when one of the two alleles was completely absent from the leiomyoma tissue or when the ratio between the allele area of the tumor tissue against normal tissue was smaller than 0.5, as described by Cawkwell et al30. 3.2.5 Statistical Analysis The Fisher’s Exact Test was used to compare the two groups of patients. The statistical analyses were done with the statistics software SigmaStat, Jandel Scientific. The significance level adopted was of 5%.
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
4.1 Microsatellite Analysis The chromatograms of the microsatellite markers of the present study (D7S471, D7S496, D7S501, D7S515, D7S518, D7S666) were obtained by automated sequencing. For some of the studied microsatellite markers, chromatogram analysis revealed same size alleles in the DNA of peripheral blood and normal tissue. These cases were considered homozygotes and noninformative (Figure 1). Cases presenting two alleles were considered heterozygotes and were informative for the study. The allele area analysis, obtained from the product of the allele peak value by its width value, as well as the resulting ratio between the tumor alleles and normal alleles revealed that, in many cases, the obtained values were higher than 0.5, therefore, in these cases, there was no loss of heterozygosity. Figure 2 shows the automated sequencing chromatograms of the D7S471 marker PCR products from peripheral blood normal genomic DNA and leiomyoma tissue DNA regarding two informative cases where there was no loss of heterozygosity. In these cases, we could also verify that the allele size in tumor tissue did not change, therefore these cases were considered stable for the studied marker. When we detected the presence of two alleles whose size differed from those of normal tissue, we considered that these cases presented MSI. Figure 3 illustrates the MSI cases. After automated sequencing, if the product of the normal DNA by tumor DNA revealed heterozygotes in normal tissue and absence of one of the tumor tissue alleles, these cases were considered LOH (Figure 4). In the same way, the cases where the tumor tissue presented two alleles similar to those of normal tissue, but the ratio between the allele areas was smaller than 0.5, these were also considered LOH. Figure 5 and Figure 6 illustrate these cases with LOH. 4.1.1 Results from the group with uterus-leiomyoma shrinkage £ 36% The ratio values between the tumor tissue and normal tissue alleles of the 06 studied microsatellites that presented a uterus-leiomyoma shrinkage equal to or below 36% after GnRH treatment are illustrated in table 3. A global analysis of the cases in this group of patients revealed a small number of cases with LOH for each one of the studied microsatellites markers. LOH occurred for all markers but in low frequencies: 20% (2/10) of LOH for the D7S471 marker, 33% (3/9) for D7S496, 22% (2/9) for D7S501, 20% (1/5) for D7S515, 11% (1/9) for D7S518 and 9% (1/11) for D7S666. These results show that, among this group of patients with a shrinkage equal to or below 36%, LOH was more frequent for the microsatellite markers that include a cluster of genes from the family DRA (down-regulated in adenoma), DLD (desidrogenase dihydrolipoamide), PRKAR2B (protein kinase, cAMP-dependent, regulatory, type II, beta) and LAMB1 (laminin, B1). The DLD gene is a maintenance gene; the DRA and the PRKAR2B are also expressed in other tissues besides the uterus, so they do not seem to be involved with uterine leiomyoma. The gene LAMB1 may somehow modify the extracellular matrix and produce a proliferation signal31 (Zeng et al 1997). We observed that the uterus-leiomyoma shrinkage £ 36% patients presented a low frequency of LOH for the D7S518, D7S666 and D7S515 microsatellites, marking genes of the CUTL1 family, that is, tumor suppressor genes31 (Zeng et al 1997). Analyzing the LOH regarding these two families of genes in chromosome 7, CUTL1 (D7S515, D7S518 and D7S666 markers) and DRA, PRKAR2B, DLD and LAMB1 (D7S471, D7S496 and D7S501) markers, we observed that this group of patients with a uterus-leiomyoma shrinkage £ 36% present a higher frequency of LOH (64%; 7/11) for genes that do not seem to be associated with tumor suppression. A small number of patients presented LOH (22%, 2/9) in the CUTL1 family markers, genes associated with tumor suppression. Analyzing the results from the group of patients with a shrinkage £ 36%, we compared the proportion of patients (2/9) that presented LOH in the microsatellite marker category associated with the CUTL1 gene (D7S515, D7S518 and D7S666) with the proportion of patients (7/11) that presented LOH in the markers associated with lowly expressive genes for leiomyoma (D7S471, D7S496 and D7S501). We verified that there is no statistically significant difference between these two categories (p=0.092) in these patients with a shrinkage £ 36%. The MSI analysis of the 06 microsatellite markers of the 7q22 chromosome revealed the presence of MSI in the marker D7S471 in 9% (1/11), D7S496 in 50% (6/12), D7S501 in 36% (4/11), D7S515 in 22% (2/9), D7S518 in 36% (4/11) and D7S666 in 33% (4/12) of the studied cases. Considering the presence of MSI in all 06 markers, 58% (7/12) of the patients in this group (shrinkage £ 36%) presented a high frequency of instability, i.e., presented MSI in 2 or more of the 06 studied markers (Table 3). 4.1.2 Results from the group with a uterus-leiomyoma shrinkage >36% The results of the association between the normal tissue and tumor tissue alleles of the 06 studied microsatellite markers of the patients who had a uterus-leiomyoma shrinkage >36% after treatment with GnRH are specified in Table 4. Table 4 shows that a high number of patients presented LOH in most of the studied microsatellites. We observed LOH in all markers, some presenting a higher frequency of cases: in D7S471, there was 13% (2/15) of LOH; in D7S496, 20% (3/15); in D7S501, 43% (6/14); in D7S515, 43% (6/14); in D7S518, 26% (4/15); and in D7S666, 40% (6/15) of LOH. In this group of patients with a shrinkage >36%, LOH was more frequent for the microsatellite marker that includes the LAMB1 gene, associated with the extracellular matrix and, more importantly, we observed a high frequency of LOH in the markers associated with the CUTL1 family, tumor suppressor genes. Analyzing the frequency of LOH regarding these two chromosome 7 family of genes, CUTL1 (D7S515, D7S518 and D7S666 markers) and markers for DRA, PRKAR2B, DLD and LAMB1 (D7S471, D7S496 and D7S501), we observed that these patients with a shrinkage >36% presented a higher frequency of LOH (75%; 12/16) in the CUTL1 family, genes associated with tumor suppression. Regarding the other family, we also observed a high frequency of LOH, 53% (9/17), resultant from the high frequency of LOH in the D7S501 marker. Analyzing the results from the group of patients with shrinkage >36%, we compared the proportion of patients (12/16) who presented LOH in the microsatellite marker category associated with the gene CUTL1 (D7S515, D7S518 and D7S666) with the proportion of patients (9/17) that presented LOH in the markers associated with lowly expressive genes for leiomyoma (D7S471, D7S496 and D7S501). We verified that there was no statistically significant difference between these two categories (p=0.282) among these patients with a shrinkage >36%. Regarding the study of MSI of the 06 chromosome 7q22 microsatellite markers, we observed the presence of MSI in all markers: D7S471 in 11% (2/17), D7S496 in 23% (4/17), D7S501 in 41% (7/17), D7S515 in 38% (6/16), D7S518 in 18% (3/17) and D7S666 in 35% (6/17). Considering the presence of MSI in all 06 markers, 53% (9/17) of the patients in this group (shrinkage >36%) presented a high frequency of instability, i.e., presented MSI in 2 or more of the 06 studied markers. Three of the 17 studied cases presented low MSI, i.e., presented MSI in only one marker of the 06 studied markers. Five cases did not present MSI and were considered stable for these microsatellite markers. 4.1.3 Comparative analysis of the results between the studied groups Analyzing the results of the presence of high MSI (two or more markers) and using the Fisher’s Exact Test to compare the proportion of patients in the group with a shrinkage £ 36% (7/12) with the patients with a shrinkage >36% (9/17), we verified that there was no statistically significant difference (p>0.05) in the MSI proportions between these two groups. Using the Fisher´s Exact test to compare the two studied groups, the proportion of patients that presented LOH in any of the 06 markers was of 9/12 patients with a uterus-leiomyoma shrinkage £ 36% and 16/17 patients with a uterus-leiomyoma shrinkage >36% after GnRH treatment. We verified that there was no statistically significant difference (p=0.279) in the LOH proportion between these groups. Comparing the proportion of patients that presented LOH in the marker category associated with the gene CUTL1 (D7S515, D7S518 and D7S666) of the group with a shrinkage £ 36% (2/9) and the group with a shrinkage >36% (12/16), we verified a statistically significant difference (p=0.017). Comparing the proportion of patients that presented LOH in the marker category associated with lowly expressive genes for leiomyoma (D7S471, D7S496 and D7S501) of the group with a shrinkage £ 36% (7/11) and the group with a shrinkage >36% (9/17), we verified that there was no statistically significant difference (p=0.705). Analyzing the proportion of patients who presented 02 or more markers with LOH and the proportion of patients that presented only one marker with LOH, we verified, using the Fisher’s Exact Test, that there is a significant difference (p=0.040) between the two studied groups. Thus, the group with a shrinkage >36% presented a higher proportion of patients with LOH in two or more markers. Table 1: Patients with reduction £ 36%
Table 2: Patients with reduction > 36%
Table 3. Patients with reduction £ 36%
Table 4. Patients with reduction >36%.
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
Studies have demonstrated that GnRH agonist administration in patients with uterine leiomyoma reduces the volume of the tumor, the ovarian hormone levels and some growth factors that are required for tumor growth14,15. Although the mechanism by which the GnRH agonist acts on uterine leiomyoma has not been clarified yet, an induction of cellular atrophy and a reduction in the number of cells, probably resultant from apoptosis, have been suggested. Evidences exist favoring cellular damage, degeneration and necrosis, which would contribute to the tumor shrinkage after GnRH analog treatment32,33. It is known that GnRH treatment reduces uterine blood flow because of an increase in vascular resistance34,35. Ischemia causes oxidative stress which damages the DNA and consequently activates the DNA repair and cell protection mechanisms36. For most cells, oxidative stress is sublethal and reversible. However, for cells which present DNA damage, the depletion of energy causes cell death. Experimental studies verified an increased expression of the nuclear enzyme associated with DNA repair: PARP (poly ADP-ribose polymerase) in response to oxidative DNA damage. This enzyme stimulates DNA repair, silences transcription for cell growth and increases ATP generation to maintain cell survival37. A prolonged PARP activation causes an elevated consumption of ATP and this promotes cell death in injured cells by energy depletion38. Huang et al.39 demonstrated an accumulation of PARP in leiomyoma when compared with the myometrium and an overexpression of PARP, especially in the central portion of the tumor, after GnRH treatment. Regarding the tumors that present DNA alteration, we could infer that leiomyomata with MSI or LOH would be more susceptible to GnRH action since hypoestrogenism promotes oxidative stress, which results in greater DNA damage. In this way, these tumors would present an overexpression of PARP in order to repair DNA, leading to a high energy consumption resulting in cell death. However, in our study, we did not verify a significant association between leiomyomata and MSI, and the amount of shrinkage. Regarding the presence of LOH in any of the 06 studied markers, we have also not observed an association with the amount of leiomyoma shrinkage after GnRH treatment. Takahashi et al25 also did not find a significant correlation between LOH and amount of uterine leiomyoma shrinkage after GnRH treatment. However, when we selected the patients who presented LOH in 03 markers associated with the tumor suppressor gene (D7S515, D7S518, D7S666), we observed a higher proportion of LOH among the shrinkage >36% patients, with a significant association between the presence of LOH in the markers for tumor suppressor genes and greater leiomyoma shrinkage after GnRH treatment.
These results concur that this 7q22 region is critical for uterine leiomyoma, where the gene CUTL1 is considered a tumor suppressor gene whose function is to suppress the protooncogene c-myc40. Other authors have also verified this fact demonstrating reduced expression of the CUTL1 gene mRNA in uterine leiomyomata31. Another category of studied microsatellites (D7S471, D7S496 and D7S501), found in the 7q22 region, include the DRA, PRKAR2B and DLD genes, which have a low association with tumor or uterine leiomioma31. Also in this 7q22 region, adjacent to the D7S501 marker, is the LAMB1 gene (laminin B1), which presents an important interaction between malignant cells and extracellular matrix41. In this context, it is not very clear how the LAMB1 gene loss of function could contribute for uterine leiomyoma cell transformation. Our results demonstrated that the patients with lower leiomyoma shrinkage after GnRH treatment, presented higher frequency of LOH in these leiomyoma lowly pathogenesis-associated markers. These data suggest that the stimulus for proliferation of these tumors can be different and lowly affected by GnRH action. When we correlate the data obtained in the present study with the data obtained in a previous study in this same group of patients2, we verified that uterine leiomyomata with greatest shrinkage presented a lower quantity of estrogen receptors. It is known that estrogen is highly promoting for these cells; therefore, cells with few estrogen receptors seem to be more susceptible to the hypoestrogenism resultant from GnRH treatment.
Cheng et al. 42 demonstrated that the suppression of growth factors, such as estrogen, causes DNA damage. When DNA is damaged, cell replication is inhibited and the DNA repair system is activated to maintain gene integrity and normal cell function.
Thus, leiomyoma shrinkage after GnRH treatment seems to derive from hypoestrogenism, which promotes DNA damage by ischemia, activating the repair mechanisms that consume a lot of energy, promoting cell death. These data agree with our findings of a higher proportion of DNA damage, revealed by the presence of LOH, especially in the markers associated with the tumor suppressor gene, among the patients that present greater leiomyoma shrinkage after GnRH treatment. In favor of this hypothesis, the patients that presented greater leiomyoma shrinkage after GnRH treatment presented LOH in 2 or more markers. Meanwhile, the patients with less shrinkage presented LOH in only 01 marker. Therefore, we found a significant association between greater DNA damage and greater leiomyoma shrinkage.
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
Uterine leiomyomata with a higher number of changes in the 7q22 chromosome or changes in the CUTL1 gene region seem to be more susceptible to GnRH action. The pathogenic basis for the varying degrees of uterine leiomyomata shrinkage after GnRH treatment seems to be associated with the number of estrogen receptors and the DNA damage of these cells.
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
The authors thank FAPESP (Fundação de Amaparo à Pesquisa do Estado de São Paulo) for the financial support (Process 01/14052-5).
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
1 - Adamson GD. Treatment of uterine fibroids: current findings with gonadotropin-releasing hormone agonists. The estrogen threshold hypothesis. Am J Obstet Gynecol 1992 Feb;166(2):746-51 2 - Bozzini N, Rodrigues CJ, Petti DA et al. Effects of treatment with gonadotropin releasing hormone agonist on the uterine leiomyomata structure. Acta Obstet Gynecol Scand 2003; 82:330-334. 3 - Rein MS. Advances in uterine leiomyoma research: the progesterone hypothesis. Environ Health Perspect 2000;108:791-793. 4 - Ross RK, Pik MC, Vessey MP et al. Risk factors for uterine fibroids: reduced risk associated with oral contraceptives. Br Med J 1986; 293:359-362. 5 - Van Voorhis BJ, Romitti PA, Jones MP. Family History as a risk factor for development of uterine leiomyomas. J Reprod Med 2002; 47: 663-669. 6 - 7 - Wilson EA, Yang F, Rees D. Estradiol and progesterone binding In uterine leiomyomata and in normal uterine tissues. Obstet Gynecol 1980; 55:20-24. 8 - Regidor PA, Schimidt M, Callies R et al. Estrogen and progesterone receptor content of GnRH analogue pretreated and untreated uterine leiomyomata. Eur J Obstet Gynecol Reprod Biol 1995; 63:69-71. 9 - Vercellini P, Bocciolone L, Colombo A et al. Gonadotropin releasing hormone agonist treatment before hysterectomy for menorrhagia and uterine leiomyomas. Acta Obstet Gynecol Scand 1993; 72:369-373. 10 - Cirkel U, Ochs H, Roehl A et al. Estrogen and progesterone receptor content of enucleated uterine myomata after luteinizing hormone-releasing hormone. Acta Obstet Gynecol Scand 1994; 73:328-332. 11 - Rein MS, Barbieri RL, Welch W et al. The concentrations of collagen-associated amino acids are higher in GnRH agonist-treated uterine myomas. Obstet Gynecol 1993; 82:901-905. 12 - Broekmans FJ. Gonadotropin-releasing hormone agonists and uterine leiomyomas. Hum Reprod 1996; 11:3-25. 13 - Friedman AJ, Barbieiri RL, Benacerraf BR et al. Treatment of leiomyomata with intranasal or subcutaneous leuprolide, a gonadotropin-releasing hormone agonist. Fertil Steril 1987; 48:560-564. 14 - Liu CH, Lin YS, Lin CC et al. Medical Treatment of uterine myoma with long acting gonodotropin-relasing hormone agonist prior to myomectomy. J Formosan Med Assoc 1993; 92:650-654. 15 - West CP, Lumsden MA, Lawson S et al. Shrinkage of uterine fibroids during therapy with goserelin (Zoladex), a aluteinizing hormone agonist administered as a monthly subcutaneous depot. Fertil Steril 1987; 48: 45-55. 16 - Friedman AJ, Rein MS, Harrison-Atlas D et al. A randomized, placebo-controlled, double blind study evaluating leuprolide acetate depot treatment prior to myomectomy. Fertil Steril 1989; 52:728-33. 17 - Kalir T, Goldstein M, Dottino P et al. Morphometric and electron-microscopic analyses of the effects of GnRH agonists on uterine leiomyomas. Arch Pathol Lab Med 1998; 122: 442-446. 18 - Boghosian L, Dal Cin P, Sandberg AA. An interstitial deletion of chromosome 7 may characterize a subgroup of uterine leiomyoma. Cancer Genet Cytogenet 1988; 34:207-208. 19 - Gibas Z, Griffin AC, Emanuel BS. Clonally chromosomal rearrangement in a uterine myoma. Cancer Genet Cytogenet 1988; 32:19-24. 20 - Heim S, Nilbert M, Vanni R et al. A specific translocation, t (12; 14)(q14-15: q23-24) characterizes a subgroup of uterine leiomyomas. Cancer Genet Cytogenet 1988; 32:13-17. 21 - Mark J, Havel G, Grepp C et al. Chromosomal patterns in human benign uterine leiomyomas. Cancer Genet Cytogenet 1990; 44:1-13. 22 - Nilbert M, Heim S, Mandahl N et al. Characteristic chromosome abnormalities, including rearrangements of 6p, del(7q), +12, and t(12;14) in 44 uterine leiomyomas. Hum Genet 1990; 85:605-611. 23 - Ozisik YY, Meloni AM, Surti U et al. Deletion 7q22 in uterine leiomyoma: A cytogenetic review. Cancer Genet Cytogenet 1993; 71:1-6. 24 - Turc-Carel C, Dal Cin P, Boghosian L et al. Consistent breakpoints in region 14q22-24 in uterine leiomyoma. Cancer Genet Cytogenet 1988; 32: 25-31. 25 - Takahashi K, Kawamura N, Tsujimura A et al. Association of the shrinkage of uterine leiomyoma treated with GnRH agonist and deletion of long arm of chromosome 7. Int J Oncol 2001; 18:1259-1263. 26 - Dib C, Faure S, Fizames C et al. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 1996; 380:152-154. 27 - Mayr W. DNA markers in forensic medicine. Transfus Clin Biol 1995; 4:325-328. 28 - Nascimento E, Spinelli M, Rodrigues CJ, Bozzini N. Protocolo de extração de DNA de material parafinado para análise de microssatélites em leiomioma uterino. J Bras Patol 2003; 39:253-255. 29 - Miller AS, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acid Res 1988; 16:1215. 30 - Cawkwell L, Bell SM, Lewis FA et al. Rapid detection of allele loss in colorectal tumours using microsatellites and fluorescent DNA technology. Br J Cancer 1993; 67:1262-1267. 31 - Zeng WR, Scherer SW, Koutsilieris M et al. Loss of heterozygosity and reduced expression of the CUTL1 gene in uterine leiomyomas. Oncogene 1997; 14:2355-2365. 32 - Deligdisch L, Hirschmann S, Altchek A. Pathologic changes in GnRH agonist analogue treated uterine leiomyomata. Fertil Steril 1997; 67: 837-841. 33 - Ito F, Kawamura N, Ichimura T et al. Ultrastructural comparison of uterine leiomyoma cells from the same myoma nodule before and after GnRH agonist treatment. Fertil Steril 2001; 75:125-130. 34 - Matta WH, Stabile I, Shaw RW et al. Doppler assessment of uterine blood flow changes in patients with fibroids receiving GnRH agonist Buserelin. Fertil Steril 1988; 46:1083-1085. 35 - Spong CY, Sinow R, Renslo R et al. Induced hypoestrogenism increases the arterial resistance index of leiomyomata without affecting uterine or carotid arteries. J Assist Reprod Genet 1995; 12:338-341. 36 - Nagayama T, Simon RP, Chen D et al. Activation of PARP in the rat hippocampus may contribute to cellular recovery following sublethal transient global ischemia. J Neurochem 2000; 74:1636-1645. 37 - Ziegler M, Oei SL. A cellular survival switch: PARP stimulates DNA repair and silences transcription. Bioessays 2001; 23:543-548. 38 - Endres M, Wang ZQ, Namura S et al. Ischemic brain injury is mediated by the activation of PARP . J Cereb Blood Flow Metab 1997; 1711:1143-1151. 39 - Huang SC, Tang MJ, Cheng YM et al. Enhanced PARP in GnRH agonist-treated uterine leiomyoma. J Clin Endocrinol Metab 2003; 88:5009-5016. 40 - Lemieux N, Zhang XX, Dufort D et al. Assignment of the human homologue of Drosophila Cut homeobox gene to band 7q22 by fluorescence in situ hybridization. Genomics 1994; 24:191-193. 41 - Honn KV, Tang DG. Cancer & Metastasis Reviews 1992; 11:353-375. 42 - Cheng YM, Chou CY, Huang SC et al. Oestrogen deficiency causes DNA damage in uterine leiomyoma cells: a possible mechanism for shrinkage of fibroids by GnRH
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|
Web mantenido y actualizado por el Servicio de informática uclm. Modificado: 16/06/2015 15:17:25
 fiogf49gjkf0dFigure 1.">
fiogf49gjkf0dFigure 1.">
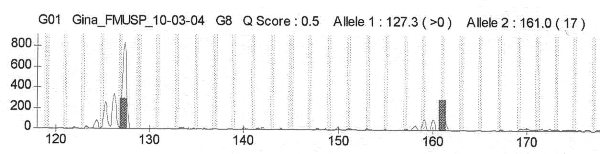
 fiogf49gjkf0dFigure 2. Cromatogram from patient G2 for D7S666. Heterozygous and have LOH">
fiogf49gjkf0dFigure 2. Cromatogram from patient G2 for D7S666. Heterozygous and have LOH">
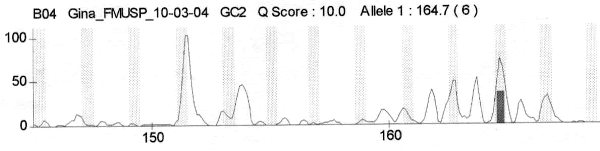 fiogf49gjkf0dFigure 3. Patient G2 - DNA leiomyoma">
fiogf49gjkf0dFigure 3. Patient G2 - DNA leiomyoma">
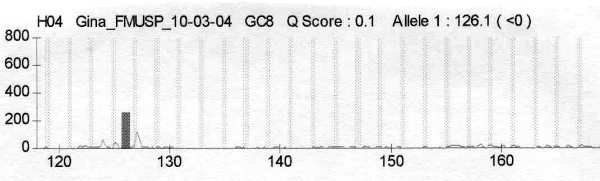
 fiogf49gjkf0dFigure 4.">
fiogf49gjkf0dFigure 4.">
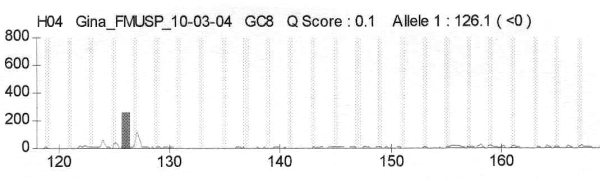 fiogf49gjkf0dFigure 5. Cromatograms from patient G8 which is homozygous for DS515">
fiogf49gjkf0dFigure 5. Cromatograms from patient G8 which is homozygous for DS515">